
Large-scale pancreatic cancer detection via non-contrast CT and deep learning
Pancreatic ductal adenocarcinoma (PDAC), the most deadly solid malignancy, is typically detected late and at an inoperable stage. Early or incidental detection is associated with prolonged survival, but screening asymptomatic individuals for PDAC using a single test remains unfeasible due to the low prevalence and potential harms of false positives. Non-contrast computed tomography (CT), routinely performed for clinical indications, ofers the potential for large-scale screening, however, identifcation of PDAC using non-contrast CT has long been considered impossible. Here, we develop a deep learning approach, pancreatic cancer detection with artifcial intelligence (PANDA), that can detect and classify pancreatic lesions with high accuracy via non-contrast CT. PANDA is trained on a dataset of 3,208 patients from a single center. PANDA achieves an area under the receiver operating characteristic curve (AUC) of 0.986–0.996 for lesion detection in a multicenter validation involving 6,239 patients across 10 centers, outperforms the mean radiologist performance by 34.1% in sensitivity and 6.3% in specifcity for PDAC identifcation, and achieves a sensitivity of 92.9% and specifcity of 99.9% for lesion detection in a real-world multi-scenario validation consisting of 20,530 consecutive patients. Notably, PANDA utilized with non-contrast CT shows non-inferiority to radiology reports (using contrast-enhanced CT) in the diferentiation of common pancreatic lesion subtypes. PANDA could potentially serve as a new tool for large-scale pancreatic cancer screening.
Effective Opportunistic Esophageal Cancer Screening using Noncontrast CT Imaging
Esophageal cancer screening using Non-contrast CT

Deep Learning for Fully Automated Prediction of Overall Survival in Patients Undergoing Resection for Pancreatic Cancer: A Retrospective Multicenter Study
Another pancreatic cancer work milestone for our group.

DeepPrognosis: Preoperative Prediction of Pancreatic Cancer Survival and Surgical Margin via Comprehensive Understanding of Dynamic Contrast-Enhanced CT Imaging and Tumor-Vascular Contact Parsing
MICCAI-MedIA (Medical Image Analysis) Special Issue of Best Papers in 2020
3D Graph Anatomy Geometry-Integrated Network for Pancreatic Mass Segmentation, Diagnosis, and Quantitative Patient Management
Nice work from our Intern. It the first work to propose a multiphase CT imaging analysis method for the full-spectrum taxonomy of pancreatic mass/disease diagnosis.

Deep learning for fully-automated prediction of overall survival in patients with oropharyngeal cancer using FDG PET imaging
My co-first authorship clinical paper on oncology.
DeepPrognosis: Preoperative Prediction of Pancreatic Cancer Survival and Surgical Margin via Contrast-Enhanced CT Imaging
A Contrast-Enhanced CT prediction model for PDAC.
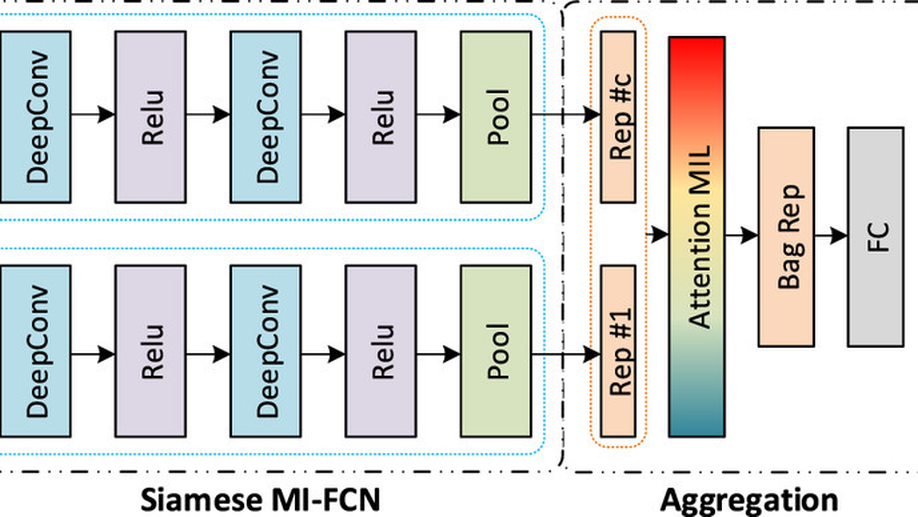
Whole Slide Images based Cancer Survival Prediction using Attention Guided Deep Multiple Instance Learning Networks
This is an improved version of our MICCAI 19 work. Most cited articles of MedIA since 2020.
An efficient algorithm for dynamic MRI using low-rank and total variation regularizations
The extension version of MICCAI 2015. MATLAB Code is available.
Wsisa: Making survival prediction from whole slide histopathological images
Use Deep learning model to predict survival from WSI.
Background subtraction based on low-rank and structured sparse decomposition
Extension of ICME 2014, MATLAB code is available.